3,5,7-Substituted Pyrazolo[4,3- d ]Pyrimidine Inhibitors of Cyclin-Dependent Kinases and Cyclin K Degraders.
Jorda, R., Havlicek, L., Perina, M., Vojackova, V., Pospisil, T., Djukic, S., Skerlova, J., Gruz, J., Renesova, N., Klener, P., Rezacova, P., Strnad, M., Krystof, V.(2022) J Med Chem 65: 8881-8896
- PubMed: 35749742
- DOI: https://doi.org/10.1021/acs.jmedchem.1c02184
- Primary Citation of Related Structures:
7QHL - PubMed Abstract:
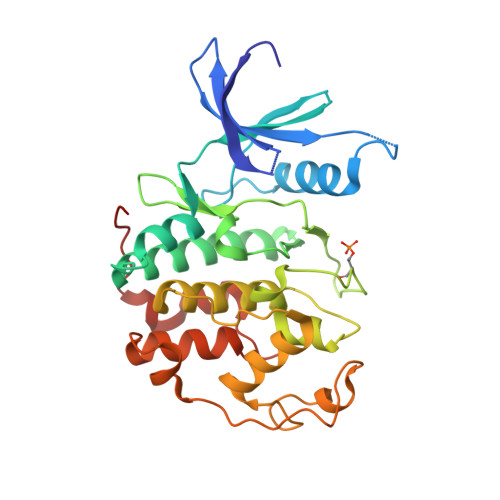
3,5,7-Trisubstituted pyrazolo[4,3- d ]pyrimidines have been identified as potent inhibitors of cyclin-dependent kinases (CDKs), which are established drug targets. Herein, we describe their further structural modifications leading to novel nanomolar inhibitors with strong antiproliferative activity. We determined the crystal structure of fully active CDK2/A2 with 5-(2-amino-1-ethyl)thio-3-cyclobutyl-7-[4-(pyrazol-1-yl)benzyl]amino-1(2) H -pyrazolo[4,3- d ]pyrimidine ( 24 ) at 1.7 Å resolution, confirming the competitive mode of inhibition. Biochemical and cellular assays in lymphoma cell lines confirmed the expected mechanism of action through dephosphorylation of retinoblastoma protein and RNA polymerase II, leading to induction of apoptosis. Importantly, we also revealed an interesting ability of compound 24 to induce proteasome-dependent degradation of cyclin K both in vitro and in a patient-derived xenograft in vivo. We propose that 24 has a dual mechanism of action, acting as a kinase inhibitor and as a molecular glue inducing an interaction between CDK12 and DDB1 that leads to polyubiquitination of cyclin K and its subsequent degradation.
- Department of Experimental Biology, Faculty of Science, Palacký University Olomouc, Šlechtitelů 27, 78371 Olomouc, Czech Republic.
Organizational Affiliation: